Introduction to Arrays
Last updated on 2024-05-23 | Edit this page
Download Chapter notebook (ipynb)
Mandatory Lesson Feedback Survey
Overview
Questions
- What are different types of arrays?
- How is data stored and retrieved from an array
- Why nested arrays?
- What are tuples?
Objectives
- Understanding difference between lists and tuples.
- Building concepts of operations on arrays.
- knowing storing multidimensional data.
- Understanding mutability and immutability.
So far, we have been using variables to store individual values. In some circumstances, we may need to access multiple values to perform operations. In such occasions, defining a variable for every single value can become very tedious. To address this, we use arrays.
Arrays are variables that hold any number of values. Python provides 3
types of built-in arrays: list, tuple, and
set. There are a several common features amongst all arrays
in Python; however, each type of array enjoys its own range of unique
features that facilitate specific operations.
Lists
Lists are the most frequently used type of arrays in Python. It is therefore important to understand how they work, and that how can we use them and features they offer to our advantage.
The easiest way to imagine how a list works is to think of
it as a table that can have any number of rows. This is akin to a
spreadsheet with one column. For instance, suppose we have a table with
4 rows in a spreadsheet application as follows:
The number of rows in an array determine the length. The above table has 4 rows; therefore it is said to have a length of 4.
Implementation
OUTPUT
[5, 21, 5, -1]OUTPUT
<class 'list'>Do it Yourself
Implement a list array called fibonacci, whose members
represent the first 8 numbers of the Fibonacci
sequence as follows:
| FIBONACCI NUMBERS (FIRST 8) | |||||||
|---|---|---|---|---|---|---|---|
| 1 | 1 | 2 | 3 | 5 | 8 | 13 | 21 |
Indexing
In arrays, an index is an integer number that corresponds to a specific item.
You can think of an index as a unique reference or a key that corresponds to a specific row in a table. We don’t always write the row number when we create a table. However, we always know that the 3rd row of a table means that we start from the first row (row #1), count 3 rows down and there we find the 3rd row.
The only difference in Python is that we don’t take the first row as row #1; instead, we consider it to be row #0. As a consequence of starting from #0, we count rows in our table down to row #2 instead of #3 to find the 3rd row. So our table may in essence be visualised as follows:
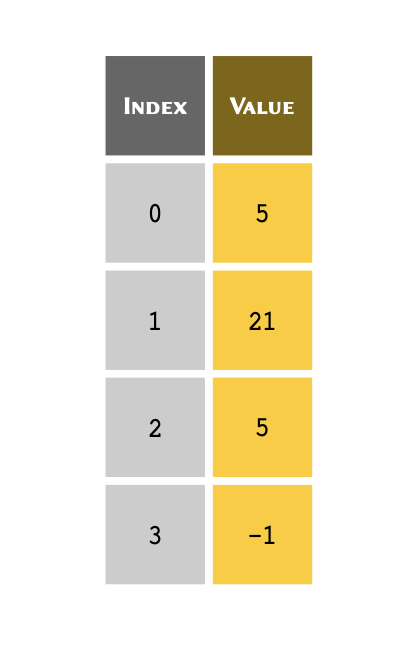
With that in mind, we can use the index for each value to retrieve it
from a list.
Given a list of 4 members stored in a variable called
table:
table = [5, 21, 5, -1]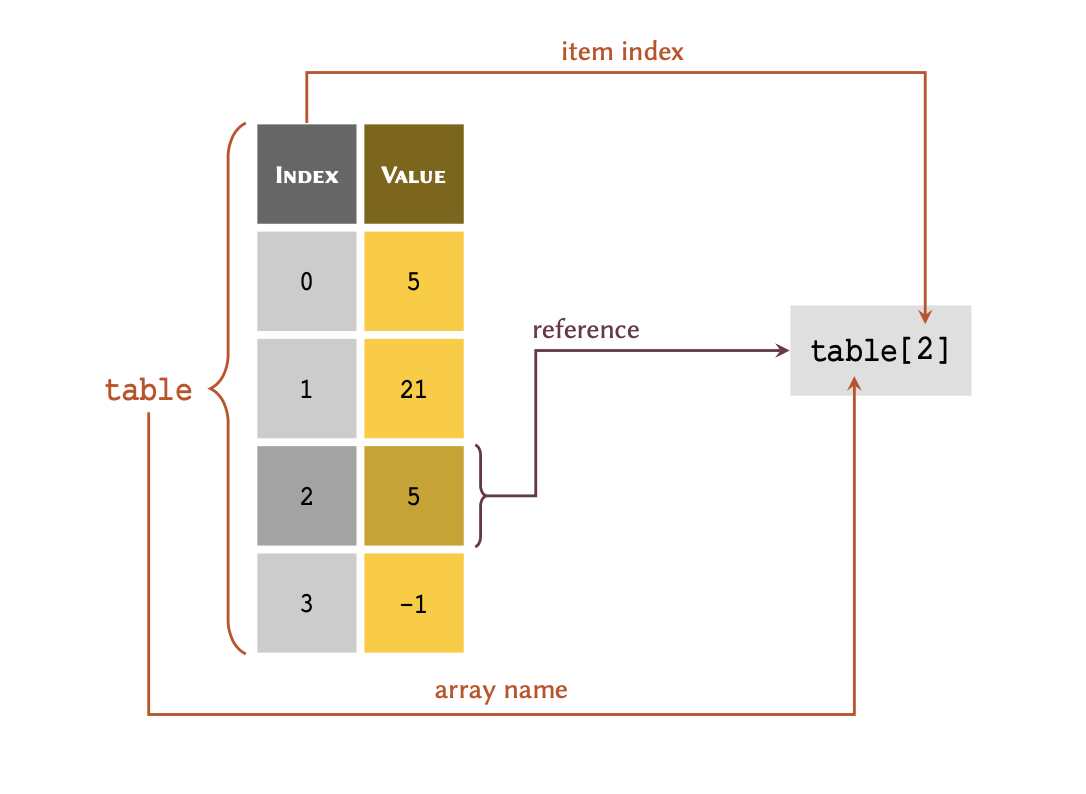
As demonstrated in the diagram; to retrieve a member of an array through its index, we write the name of the variable immediately followed by the index value inside a pair of square brackets — e.g. table[2].
OUTPUT
5OUTPUT
5OUTPUT
-1Do it Yourself
Retrieve and display the 5th Fibonacci number from the
list you created in previous DIY.
It is sometimes more convenient to index an array backwards — that is, to reference the members from the bottom of the array. This is called negative indexing and is particularly useful when we are dealing with very lengthy arrays. The indexing system in Python support both positive and negative indexing systems.
The table above therefore may also be represented as follows:
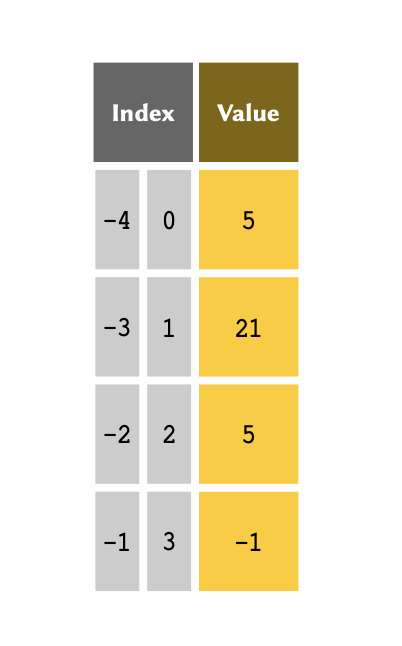
If the index is a negative number, the indices are counted from the
end of the list. We can implement negative indices the same
way we do positive ones:
OUTPUT
-1OUTPUT
5OUTPUT
21We know that in table, index #-3 refers the same value as index #1. So let us go ahead and test this:
OUTPUT
TrueIf the index requested is larger than the length of the
list minus one, an IndexError will be
raised:
OUTPUT
IndexError: list index out of rangeDo it Yourself
Retrieve and display the last Fibonacci number from the
list you created in DIY.
Slicing
We may retrieve more than one value from a list at a
time, as long as the values are in consecutive rows. This
process is known as , and may be visualised as follows:
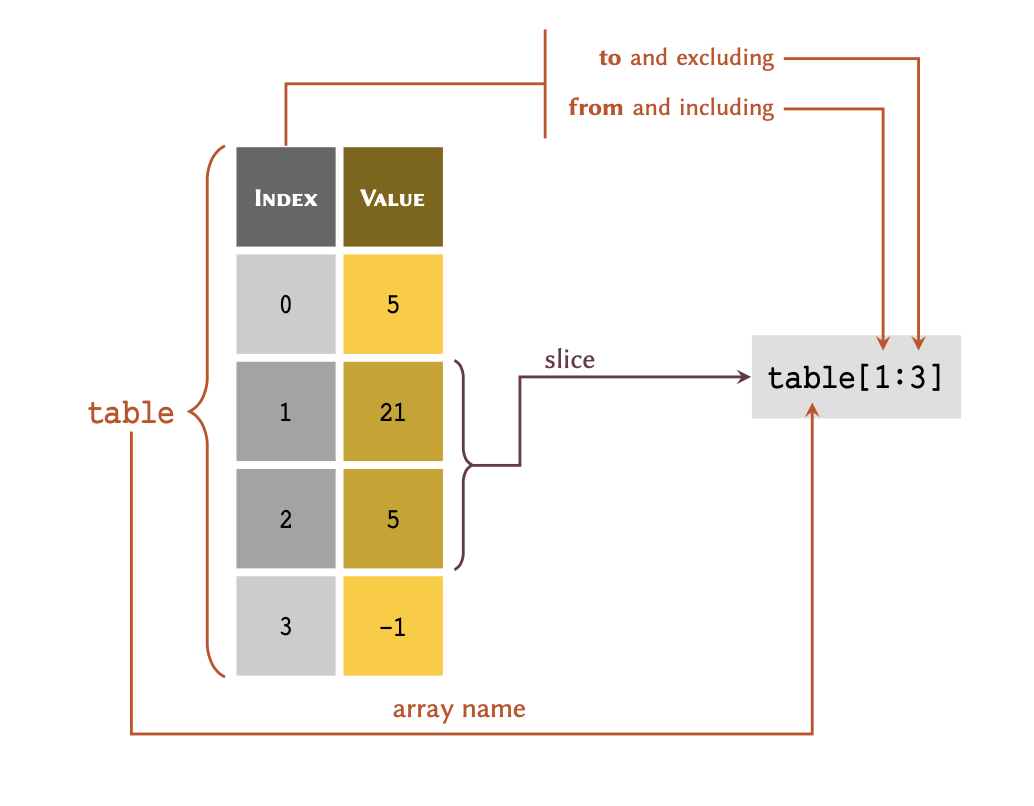
Given a list representing the above table:
table = [5, 21, 5, -1]we may retrieve a slice of table as follows:
OUTPUT
[21, 5]print(table[0:2])If the first index of a slice is #0, the slice may also be written as:
OUTPUT
[5, 21]Negative slicing is also possible:
PYTHON
# Retrieves every item from the first member down
# to, but excluding the last one:
print(table[:-1])OUTPUT
[5, 21, 5]OUTPUT
[21]If the second index of a slice represents the last index of a
list, it be written as:
OUTPUT
[5, -1]OUTPUT
[21, 5, -1]We may store indices and slices in variables:
OUTPUT
[21, 5]The slice() function may also be used to create a slice variable:
OUTPUT
[21, 5]Do it Yourself
Retrieve and display a slice of Fibonacci numbers from the
list you created in DIY that includes all the members
from the 2nd number onwards — i.e. the slice must not include
the first value in the list.
Note
Methods are features of Object-Oriented
Programming (OOP), a programming paradigm that we do not discuss in
the context of this course. You can think of a method as a
function that is associated with a specific type. The
job of a method is to provide a certain functionality unique to
the type it is associated with. In this case,
.index() is a method of type list
that given a value, finds and produces its index from the
list.
From value to index
Given a list entitled table as:
we can also find out the index of a specific value. To do so, we use the .index() method:
OUTPUT
1OUTPUT
3If a value is repeated more than once in the list, the
index corresponding to the first instance of that value is
returned:
OUTPUT
0If a value does not exist in the list, using
.index() will raise a ValueError:
OUTPUT
ValueError: 9 is not in listDo it Yourself
Find and display the index of these values from the list
of Fibonacci numbers that you created in DIY:
- 1
- 5
- 21
Mutability
Arrays of type list are modifiable. That is, we can add new
values, change the existing ones, or remove them from the array all
together. Variable types that allow their contents to be modified are
referred to as mutable types in programming.
Addition of new members
Given a list called table as:
We can add new values to table using .append():
OUTPUT
[5, 21, 5, -1, 29]OUTPUT
[5, 21, 5, -1, 29, 'a text']
Sometimes, it may be necessary to insert a value at a specific index in
a list. To do so, we may use .insert(), which
takes two input arguments; the first representing the index, and the
second the value to be inserted:
OUTPUT
[5, 21, 5, 56, -1, 29, 'a text']Do it Yourself
Given fibonacci the
list representing the first 8 numbers in the Fibonacci
sequence that you created in DIY:
The 10th number in the Fibonacci sequence is 55. Add this value to fibonacci.
Now that you have added 55 to the
list, it no longer provides a correct representation of the Fibonacci sequence. Alter fibonacci and insert the missing number such that your it correctly represents the first 10 numbers in the Fibonacci sequence, as follows:
| FIBONACCI NUMBERS (FIRST 8) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 2 | 3 | 5 | 8 | 13 | 21 | 34 | 55 |
Modification of members
Given a list as:
We can also modify the exiting value or values inside a
list. This process is sometimes referred to as item
assignment:
OUTPUT
[5, 174, 5, 56, -1, 29, 'a text']OUTPUT
[5, 174, 5, 19, -1, 29, 'a text']It is also possible to perform item assignment over a slice containing any number of values. Note that when modifying a slice, the replacement values must be the same length as the slice we are trying to replace:
PYTHON
print('Before:', table)
replacement = [-38, 0]
print('Replacement length:', len(replacement))
print('Replacement length:', len(table[2:4]))
# The replacement process:
table[2:4] = replacement
print('After:', table)OUTPUT
Before: [5, 174, 5, 19, -1, 29, 'a text']
Replacement length: 2
Replacement length: 2
After: [5, 174, -38, 0, -1, 29, 'a text']OUTPUT
[5, 174, 12, 0, -1, 29, 'a text']Do it Yourself
Given a list containing the first 10 prime numbers
as:
primes = [2, 3, 5, 11, 7, 13, 17, 19, 23, 29]
However, values 11 and 7 have been misplaced in the sequence. Correct the order by replacing the slice of primes that represents [11, 7] with [7, 11].
Removal of members
When removing a value from a list array, we have two
options depending on our needs: we either remove the member and retain
the value in another variable, or we remove it and dispose of the value.
To remove a value from a list without retaining it, we use
.remove(). The method takes one input argument, which is the
value we would like to remove from our list:
OUTPUT
[5, 12, 0, -1, 29, 'a text']Alternatively, we can use del; a Python syntax that we can use in this context to delete a specific member using its index:
OUTPUT
[5, 12, 0, -1, 29]As established above, we can also delete a member and retain its value. Of course we can do so by holding the value inside another variable before deleting it.
Whilst that is a valid approach, Python’s list provide us
with .pop() to simplify the process even further. The method
takes one input argument for the index of the member to be removed. It
removes the member from the list and returns its value, so
that we can retain it in a variable:
OUTPUT
Removed value: 0
[5, 12, -1, 29]Do it Yourself
We know that the nucleotides of DNA include A, C, T, and G.
Given a list representing the nucleotides of a DNA
strand as:
strand = ['A', 'C', 'G', 'G', 'C', 'M', 'T', 'A']Find the index of the invalid nucleotide in strand.
Use the index you found to remove the invalid nucleotide from strand and retain the value in another variable. Display the result as:
Removed from the strand: X
New strand: [X, X, X, ...]- What do you think happens once we run the following code, and why? What would be the final result displayed on the screen?
strand.remove('G')
print(strand)One of the two G nucleotides, the one at index 2 of the original array, is removed. This means that the .remove() method removes only first instance of a member in an array. The output would therefore be:
['A', 'C', 'G', 'C', 'M', 'T', 'A']Method–mediated operations
We already know that methods are akin to functions that are associated with a specific type. In this subsection, we will be looking into how operations are performed using methods. To that end, we will not be introducing anything new, but recapitulate what we already know from different perspectives.
So far in this chapter, we have learned how to perform different
operations on list arrays in Python. You may have noticed
that some operations return a result that we can store in a variable,
whilst others change the original value.
With that in mind, we can divide operations performed using methods into two general categories:
- Operations that return a result without changing the original array:
OUTPUT
2
[1, 2, 3, 4]- Operations that use specific methods to change the original array, but do not necessarily return anything (in-place operations):
OUTPUT
[1, 2, 3, 4, 5]If we attempt to store the output of an operation that does not a
return result inside a variable, the variable will be created, but its
value will be set to None:
OUTPUT
None
[1, 2, 3, 4, 5, 6]It is important to know the difference between these types of operations. So as a rule of thumb, when we use methods to perform an operation, we can only change the original value if it is an instance of a mutable type. See Table to find out which built-in types are mutable in Python.
The methods that are associated with immutable objects always return the results and do not provide the ability to alter the original value:
- In-place operation on a mutable object of type
list:
OUTPUT
[5, 7]- In-place operation on an immutable object of type
str:
OUTPUT
AttributeError: 'str' object has no attribute 'remove'OUTPUT
567- Normal operation on a mutable object of type
list:
OUTPUT
1- Normal operation on a mutable object of type
list:
OUTPUT
1List members
A list is a collection of members that are independent of
each other. Each member has its own type, and is therefore subject
to the properties and limitation of that type:
OUTPUT
<class 'int'>
<class 'float'>
<class 'str'>
For instance, mathematical operations may be considered a feature of all
numeric types demonstrated in Table. However, unless
in specific circumstance described in subsection Non-numeric
values, such operations do not apply to instance of type
str.
OUTPUT
[2, 2.1, 'abcdef']
Likewise, the list plays the role of a container that may
incorporate any number of values. Thus far, we have learned how to
handle individual members of a list. In this subsection, we
will be looking at several techniques that help us address different
circumstances where we look at a list from a ‘wholist’
perspective; that is, a container whose members are unknown to us.
Membership test
Membership test operations [advanced]
We can check to see whether or not a specific value is a member of a
list using the operator syntax in:
OUTPUT
TrueOUTPUT
FalseThe results may be stored in a variable:
OUTPUT
TrueSimilar to any other logical expression, we can negate membership tests by using :
OUTPUT
TrueOUTPUT
FalseFor numeric values, int and float
may be used interchangeably:
OUTPUT
TrueOUTPUT
TrueSimilar to other logical expression, membership tests may be incorporated into conditional statements:
OUTPUT
Hello JohnDo it Yourself
Given a list of randomly generated peptide sequences
as:
PYTHON
peptides = [
'FAEKE', 'DMSGG', 'CMGFT', 'HVEFW', 'DCYFH', 'RDFDM', 'RTYRA',
'PVTEQ', 'WITFR', 'SWANQ', 'PFELC', 'KSANR', 'EQKVL', 'SYALD',
'FPNCF', 'SCDYK', 'MFRST', 'KFMII', 'NFYQC', 'LVKVR', 'PQKTF',
'LTWFQ', 'EFAYE', 'GPCCQ', 'VFDYF', 'RYSAY', 'CCTCG', 'ECFMY',
'CPNLY', 'CSMFW', 'NNVSR', 'SLNKF', 'CGRHC', 'LCQCS', 'AVERE',
'MDKHQ', 'YHKTQ', 'HVRWD', 'YNFQW', 'MGCLY', 'CQCCL', 'ACQCL'
]Determine whether or not each of the following sequences exist in peptides; and if so, what is their corresponding index:
- IVADH
- CMGFT
- DKAKL
- THGYP
- NNVSR
Display the results in the following format:
Sequence XXXXX was found at index XXLength
Built-in functions: len
The number of members contained within a list defines its
length. Similar to the length of str values as discussed in
mathematical operations DIY-I and DIY-IV, we use
the built-in function len() also to determine the length of a
list:
OUTPUT
5OUTPUT
3
The len() function always returns an integer value
(int) equal to or greater than zero. We can store the
length in a variable and use it in different mathematical or logical
operations:
PYTHON
table = [1, 2, 3, 4]
items_length = len(items)
table_length = len(table)
print(items_length + table_length)OUTPUT
9OUTPUT
TrueWe can also use the length of an array in conditional statements:
PYTHON
students = ['Julia', 'John', 'Jane', 'Jack']
present = ['Julia', 'John', 'Jane', 'Jack', 'Janet']
if len(present) == len(students):
print('All the students are here.')
else:
print('One or more students are not here yet.')OUTPUT
One or more students are not here yet.Remember
Both in and len() may be used in reference to any type of array or sequence in Python.
See Table to find out which built-in types in Python are regarded as a sequence.
Do it Yourself
Given the list of random peptides defined in DIY:
- Define a
listcalled overlaps containing the sequences whose presence in peptides you confirmed in DIY. - Determine the length of peptides.
- Determine the length of overlaps.
Display yours results as follows:
overlaps = ['XXXXX', 'XXXXX', ...]
Length of peptides: XX
Length of overlaps: XXPYTHON
overlaps = list()
sequence = "IVADH"
if sequence in peptides:
overlaps.append(sequence)
sequence = "CMGFT"
if sequence in peptides:
overlaps.append(sequence)
sequence = "DKAKL"
if sequence in peptides:
overlaps.append(sequence)
sequence = "THGYP"
if sequence in peptides:
overlaps.append(sequence)
sequence = "NNVSR"
if sequence in peptides:
overlaps.append(sequence)
print('overlaps:', overlaps)OUTPUT
overlaps: ['CMGFT', 'NNVSR']Weak References and Copies
In our discussion on mutability, we also discussed some
of the in-place operations such as .remove() and
.append() that we use to modify an existing
list. The use of these operations gives rise the following
question: What if we need to perform an in-place operation, but also
want to preserve the original array?
In such cases, we create a deep copy of the original array before we call the method and perform the operation.
Suppose we have:
A weak reference for table_a, also referred to as an alias or a symbolic link, may be defined as follows:
OUTPUT
[1, 2, 3, 4] [1, 2, 3, 4]Now if we perform an in-place operation on only one of the two variables (the original or the alias) as follows:
we will in effect change both of them:
OUTPUT
[1, 2, 3, 4, 5] [1, 2, 3, 4, 5]This is useful if we need to change the name of a variable under certain conditions to make our code more explicit and readable; however, it does nothing to preserve an actual copy of the original data.
To retain a copy of the original array, however, we must perform a deep copy as follows:
OUTPUT
[1, 2, 3, 4, 5] [1, 2, 3, 4, 5]where table_c represents a deep copy of table_b.
In this instance, performing an in-place operation on one variable would not have any impacts on the other one:
OUTPUT
[1, 2, 3, 4, 5, 6] [1, 2, 3, 4, 5, 6] [1, 2, 3, 4, 5]where both the original array and its weak reference (table_a and table_b) changed without influencing the deep copy (table_c).
There is also a shorthand for the .copy() method to create
a deep copy. As far as arrays of type list are
concerned, writing:
new_table = original_table[:]is exactly the same as writing:
new_table = original_table.copy()Here is an example:
PYTHON
table_a = ['a', 3, 'b']
table_b = table_a
table_c = table_a.copy()
table_d = table_a[:]
table_a[1] = 5
print(table_a, table_b, table_c, table_d)OUTPUT
['a', 5, 'b'] ['a', 5, 'b'] ['a', 3, 'b'] ['a', 3, 'b']Whilst both the original array and its weak reference (table_a and table_b) changed in this example; the deep copies (table_c and table_d) have remained unchanged.
Do it Yourself
When defining a consensus sequence, it is common to include annotations to represent ambiguous amino acids. Four such annotations are as follows:
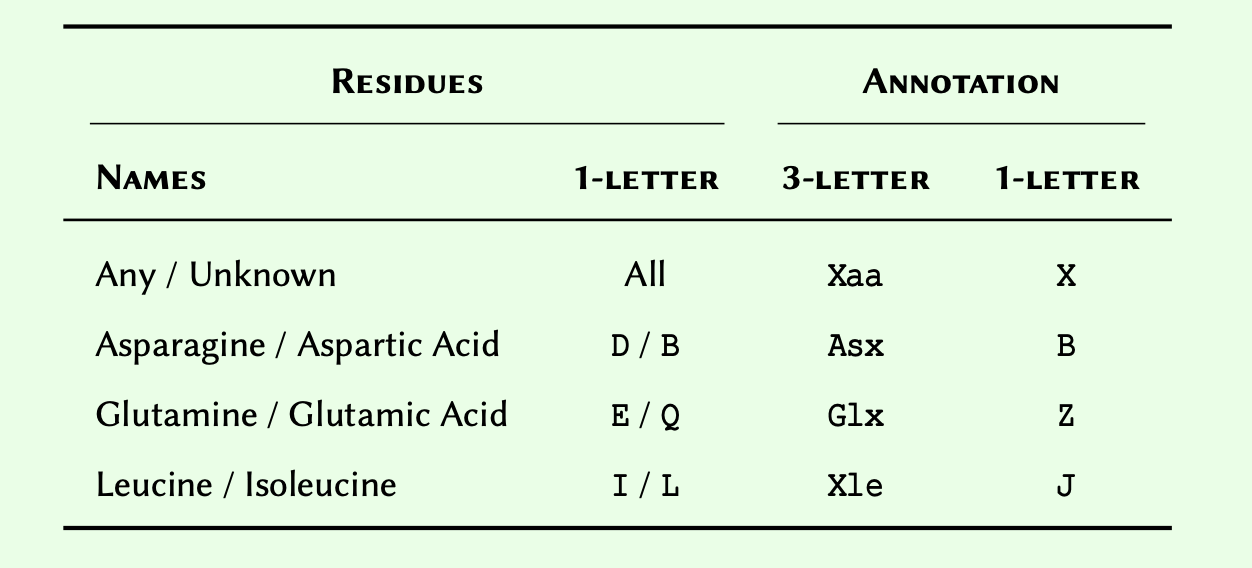
Given a list of amino acids as:
PYTHON
amino_acids = [
'A', 'R', 'N', 'D', 'C', 'E', 'Q', 'G', 'H', 'I',
'L', 'K', 'M', 'F', 'P', 'S', 'T', 'W', 'Y', 'V'
]
- Use amino_acids to
create an independent
listcalled amino_acids_annotations that contains all the standard amino acids.
- Add to amino_acids_annotations the 1-letter annotations for the ambiguous amino acids as outlined in the table.
- Evaluate the lengths for amino_acids and amino_acids_annotations and
retain the result in a new
listcalled lengths.
- Using logical
operations, test the two values stored in lengths for equivalence and
display the result as a boolean (i.e.
TrueorFalse) output.
Conversion to list
As highlighted earlier in the section, arrays in Python can contain any value regardless of type. We can exploit this feature to extract some interesting information about the data we store in an array.
To that end, we can convert any sequence
to a list. See Table to find out which
of the built-in types in Python are considered to be a sequence.
Suppose we have the sequence for Protein Kinase A Gamma (catalytic) subunit for humans as follows:
PYTHON
# Multiple lines of text may be split into
# several lines inside parenthesis:
human_pka_gamma = (
'MAAPAAATAMGNAPAKKDTEQEESVNEFLAKARGDFLYRWGNPAQNTASSDQFERLRTLGMGSFGRVMLV'
'RHQETGGHYAMKILNKQKVVKMKQVEHILNEKRILQAIDFPFLVKLQFSFKDNSYLYLVMEYVPGGEMFS'
'RLQRVGRFSEPHACFYAAQVVLAVQYLHSLDLIHRDLKPENLLIDQQGYLQVTDFGFAKRVKGRTWTLCG'
'TPEYLAPEIILSKGYNKAVDWWALGVLIYEMAVGFPPFYADQPIQIYEKIVSGRVRFPSKLSSDLKDLLR'
'SLLQVDLTKRFGNLRNGVGDIKNHKWFATTSWIAIYEKKVEAPFIPKYTGPGDASNFDDYEEEELRISIN'
'EKCAKEFSEF'
)
print(type(human_pka_gamma))OUTPUT
<class 'str'>We can now convert our sequence from its original type of
str to list by using list() as a
function. Doing so will automatically decompose the text down
to individual characters:
PYTHON
# The function "list" may be used to convert string
# variables into a list of characters:
pka_list = list(human_pka_gamma)
print(pka_list)OUTPUT
['M', 'A', 'A', 'P', 'A', 'A', 'A', 'T', 'A', 'M', 'G', 'N', 'A', 'P', 'A', 'K', 'K', 'D', 'T', 'E', 'Q', 'E', 'E', 'S', 'V', 'N', 'E', 'F', 'L', 'A', 'K', 'A', 'R', 'G', 'D', 'F', 'L', 'Y', 'R', 'W', 'G', 'N', 'P', 'A', 'Q', 'N', 'T', 'A', 'S', 'S', 'D', 'Q', 'F', 'E', 'R', 'L', 'R', 'T', 'L', 'G', 'M', 'G', 'S', 'F', 'G', 'R', 'V', 'M', 'L', 'V', 'R', 'H', 'Q', 'E', 'T', 'G', 'G', 'H', 'Y', 'A', 'M', 'K', 'I', 'L', 'N', 'K', 'Q', 'K', 'V', 'V', 'K', 'M', 'K', 'Q', 'V', 'E', 'H', 'I', 'L', 'N', 'E', 'K', 'R', 'I', 'L', 'Q', 'A', 'I', 'D', 'F', 'P', 'F', 'L', 'V', 'K', 'L', 'Q', 'F', 'S', 'F', 'K', 'D', 'N', 'S', 'Y', 'L', 'Y', 'L', 'V', 'M', 'E', 'Y', 'V', 'P', 'G', 'G', 'E', 'M', 'F', 'S', 'R', 'L', 'Q', 'R', 'V', 'G', 'R', 'F', 'S', 'E', 'P', 'H', 'A', 'C', 'F', 'Y', 'A', 'A', 'Q', 'V', 'V', 'L', 'A', 'V', 'Q', 'Y', 'L', 'H', 'S', 'L', 'D', 'L', 'I', 'H', 'R', 'D', 'L', 'K', 'P', 'E', 'N', 'L', 'L', 'I', 'D', 'Q', 'Q', 'G', 'Y', 'L', 'Q', 'V', 'T', 'D', 'F', 'G', 'F', 'A', 'K', 'R', 'V', 'K', 'G', 'R', 'T', 'W', 'T', 'L', 'C', 'G', 'T', 'P', 'E', 'Y', 'L', 'A', 'P', 'E', 'I', 'I', 'L', 'S', 'K', 'G', 'Y', 'N', 'K', 'A', 'V', 'D', 'W', 'W', 'A', 'L', 'G', 'V', 'L', 'I', 'Y', 'E', 'M', 'A', 'V', 'G', 'F', 'P', 'P', 'F', 'Y', 'A', 'D', 'Q', 'P', 'I', 'Q', 'I', 'Y', 'E', 'K', 'I', 'V', 'S', 'G', 'R', 'V', 'R', 'F', 'P', 'S', 'K', 'L', 'S', 'S', 'D', 'L', 'K', 'D', 'L', 'L', 'R', 'S', 'L', 'L', 'Q', 'V', 'D', 'L', 'T', 'K', 'R', 'F', 'G', 'N', 'L', 'R', 'N', 'G', 'V', 'G', 'D', 'I', 'K', 'N', 'H', 'K', 'W', 'F', 'A', 'T', 'T', 'S', 'W', 'I', 'A', 'I', 'Y', 'E', 'K', 'K', 'V', 'E', 'A', 'P', 'F', 'I', 'P', 'K', 'Y', 'T', 'G', 'P', 'G', 'D', 'A', 'S', 'N', 'F', 'D', 'D', 'Y', 'E', 'E', 'E', 'E', 'L', 'R', 'I', 'S', 'I', 'N', 'E', 'K', 'C', 'A', 'K', 'E', 'F', 'S', 'E', 'F']Do it Yourself
Ask the user to enter a sequence of single-letter amino acids in
lower case. Convert the sequence to list and:
- Count the number of serine and threonine residues and display the result in the following format:
Total number of serine residues: XX
Total number of threonine residues: XX- Check whether or not the sequence contains both serine and threonine residues:
- If it does, display:
The sequence does contain both serine and threonine residues.- if it does not, display:
The sequence does not contain both serine and threonine residues.sequence_str = input('Please enter a sequence of signle-letter amino acids in lower-case: ')
sequence = list(sequence_str)
ser_count = sequence.count('s')
thr_count = sequence.count('t')
print('Total number of serine residues:', ser_count)
print('Total number of threonine residues:', thr_count)if ser_count > 0 and thr_count > 0:
response_state = ''
else:
response_state = 'not'
print(
'The sequence does',
'response_state',
'contain both serine and threonine residues.'
)Advanced Topic
Generators represent a specific type in Python whose results are not immediately evaluated. This is a technique referred to as lazy evaluation in functional programming, and is often used in the context of a for-loop. This is because they postpone the evaluation of their results for as long as possible. We do not discuss generators in the course, but you can find out more about them in the official documentations.
Useful methods
Data Structures: More on Lists
In this subsection, we will be reviewing some of the useful and
important methods that are associated with object of type
list. To that end, we shall use snippets of code that
exemplify such methods in practice. A cheatsheet of the methods associated
with the built-in arrays in Python can be helpful.

The methods outline here are not individually described; however, at this point, you should be able to work out what they do by looking at their names and respective examples.
Count a specific value within a list:
OUTPUT
3Extend a list:
PYTHON
table_a = [1, 2, 2, 2]
table_b = [15, 16]
table_c = table_a.copy() # deep copy.
table_c.extend(table_b)
print(table_a, table_b, table_c)OUTPUT
[1, 2, 2, 2] [15, 16] [1, 2, 2, 2, 15, 16]Extend a list by adding two lists to each other. Note
that adding two lists is not an in-place operation:
PYTHON
table_a = [1, 2, 2, 2]
table_b = [15, 16]
table_c = table_a + table_b
print(table_a, table_b, table_c)OUTPUT
[1, 2, 2, 2] [15, 16] [1, 2, 2, 2, 15, 16]PYTHON
table_a = [1, 2, 2, 2]
table_b = [15, 16]
table_c = table_a.copy() # deep copy.
table_d = table_a + table_b
print(table_c == table_d)OUTPUT
False
We can also reverse the values in a list. There are two
methods for doing so:Being a generator means that the output of the
function is not evaluated immediately; and instead, we get a generic
output:
- Through an in-place operation using .reverse()
OUTPUT
Reversed: [16, 15, 2, 2, 2, 1]- Through reversed(), which is a build-in generator function.
PYTHON
table = [1, 2, 2, 2, 15, 16]
table_rev = reversed(table)
print("Result:", table_rev)
print("Type:", type(table_rev))OUTPUT
Result: <list_reverseiterator object at 0x7f858532f790>
Type: <class 'list_reverseiterator'>We can, however, force the evaluation process by converting the
generator results onto a list:
OUTPUT
Evaluated: [16, 15, 2, 2, 2, 1]Members of a list may be sorted in-place as follows:
OUTPUT
Sorted (ascending): [1, 2, 2, 2, 15, 16]Advanced Topic
There is also the built-in function sorted() that works in a similar way to reversed(). Also a generator function, it offers more advanced features that are beyond the scope of this course. You can find out more about it from the official documentations and examples.
The .sort() method takes an optional keyword argument
entitled reverse (default: False). If set to
True, the method will perform a descending sort:
OUTPUT
Sorted (descending): [16, 15, 2, 2, 2, 1]We can also create an empty list, so that we can add
members to it later in our code using .append() or
.extend() amongst other tools:
OUTPUT
[]OUTPUT
[5]OUTPUT
['Jane', 'Janette']This DIY was intended to encourage you to experiment with the methods outlined.
Nested arrays
At this point, you should be comfortable with creating, handling, and
manipulating arrays of type list in Python. It is important
to have a relatively good understanding of the principles outlined in
this section so far before you start learning about nested
arrays.
We have already established that arrays can contain any value regardless of type. This means that they also contain other arrays. An array that includes at least one member that is itself an array is referred to as a nested arrays. This can be thought of as a table with more than one column:
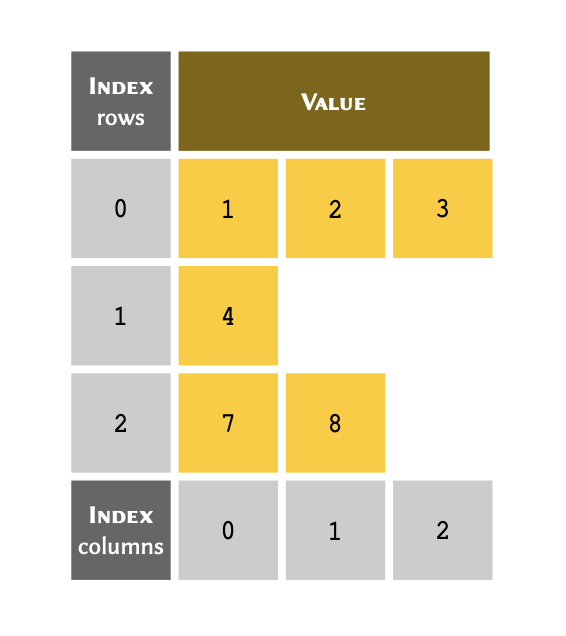
Implementation
The table can be written in Python as a nested array:
PYTHON
# The list has 3 members, 2 of which
# are arrays of type list:
table = [[1, 2, 3], 4, [7, 8]]
print(table)OUTPUT
[[1, 2, 3], 4, [7, 8]]Indexing
The indexing principles for nested arrays is slightly different. To
retrieve an individual member in a nested list, we always
reference the row index, followed by the column
index.
We may visualise the process as follows:
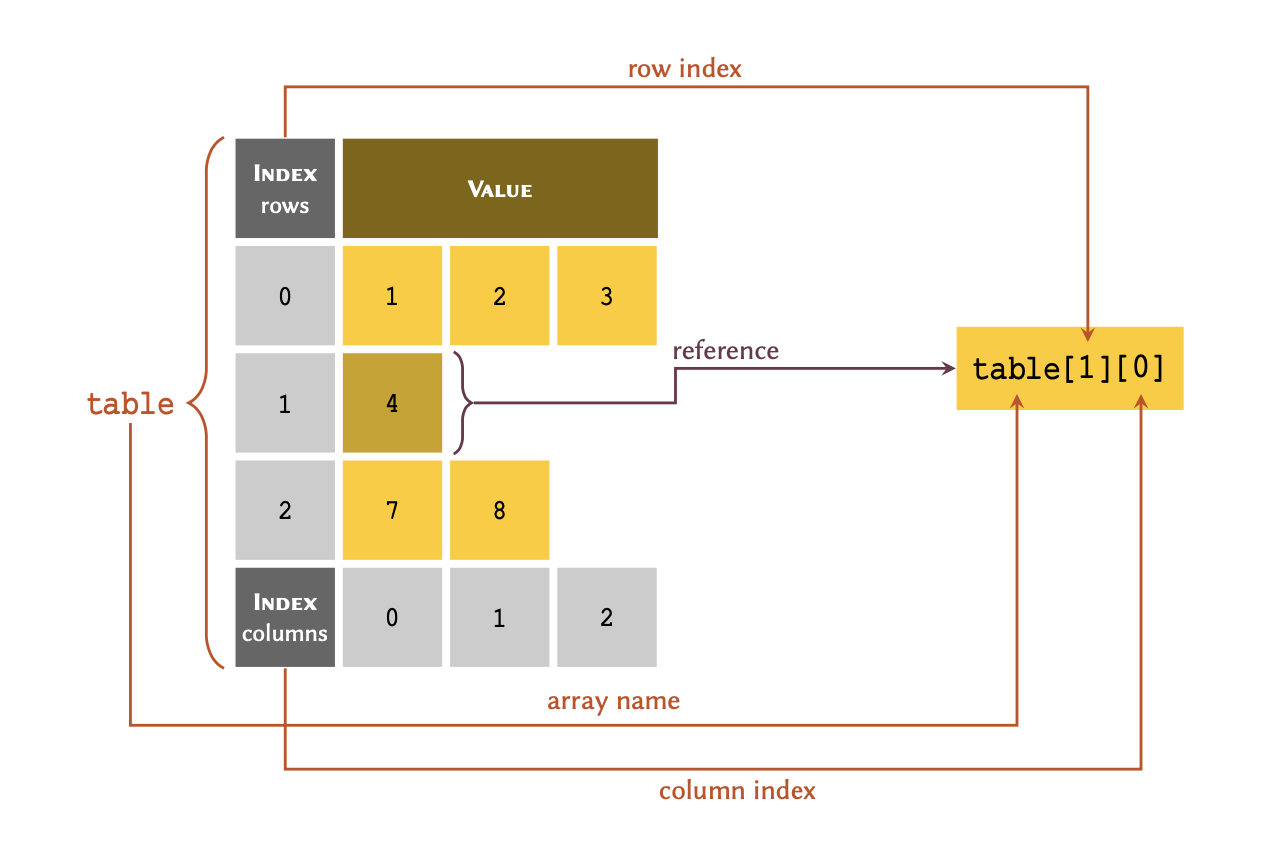
To retrieve an entire row, we only need to include the reference for that row:
OUTPUT
[1, 2, 3]and to retrieve a specific member, we include the reference for both the row and column:
OUTPUT
2We may also extract slices from a nested array. The protocol is identical to normal arrays described in subsection slicing. In nested arrays, however, we may take slices from the columns as well as the rows:
OUTPUT
[[1, 2, 3], 4]OUTPUT
[1, 2]Note that only 2 of the 3 members in table are arrays of type
list:
OUTPUT
[1, 2, 3] <class 'list'>OUTPUT
[7, 8] <class 'list'>However, there is another member that is not an array:
OUTPUT
4 <class 'int'>In most circumstances, we would want all the members in an array to be homogeneous in type — i.e. we want them all to have the same type. In such cases, we can implement the table as:
OUTPUT
[4] <class 'list'>An array with only one member — e.g. [4], is sometimes referred to as a singleton array.
Do it Yourself
Give then following of pathogens and their corresponding diseases:
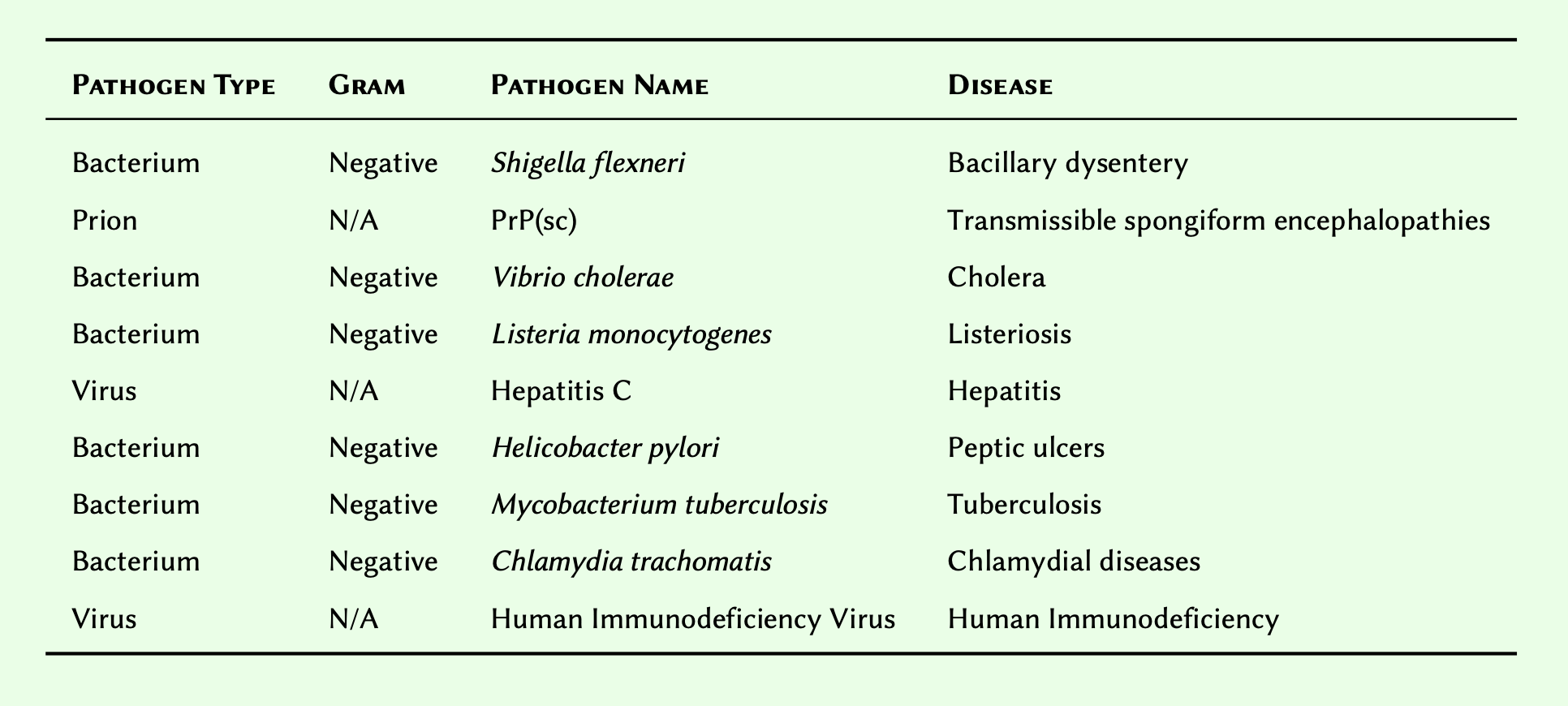
- Substituting N/A for
None, create an array to represent the table in the original order. Retain the array in a variable and display the result.
- Modify the array you created so that the members are sorted descendingly and display the result.
PYTHON
disease_pathogen = [
["Bacterium", "Negative", "Shigella flexneri" , "Bacillary dysentery"],
["Prion", None, "PrP(sc)", "Transmissible spongiform encephalopathies"],
["Bacterium", "Negative", "Vibrio cholerae", "Cholera"],
["Bacterium", "Negative", "Listeria monocytogenes", "Listeriosis"],
["Virus", None, "Hepatitis C", "Hepatitis"],
["Bacterium", "Negative", "Helicobacter pylori", "Peptic ulcers"],
["Bacterium", "Negative", "Mycobacterium tuberculosis", "Tuberculosis"],
["Bacterium", "Negative", "Chlamydia trachomatis", "Chlamydial diseases"],
["Virus", None, "Human Immunodeficiency Virus", "Human Immunodeficiency"]
]
print(disease_pathogen)OUTPUT
[['Bacterium', 'Negative', 'Shigella flexneri', 'Bacillary dysentery'], ['Prion', None, 'PrP(sc)', 'Transmissible spongiform encephalopathies'], ['Bacterium', 'Negative', 'Vibrio cholerae', 'Cholera'], ['Bacterium', 'Negative', 'Listeria monocytogenes', 'Listeriosis'], ['Virus', None, 'Hepatitis C', 'Hepatitis'], ['Bacterium', 'Negative', 'Helicobacter pylori', 'Peptic ulcers'], ['Bacterium', 'Negative', 'Mycobacterium tuberculosis', 'Tuberculosis'], ['Bacterium', 'Negative', 'Chlamydia trachomatis', 'Chlamydial diseases'], ['Virus', None, 'Human Immunodeficiency Virus', 'Human Immunodeficiency']]OUTPUT
[['Virus', None, 'Human Immunodeficiency Virus', 'Human Immunodeficiency'], ['Virus', None, 'Hepatitis C', 'Hepatitis'], ['Prion', None, 'PrP(sc)', 'Transmissible spongiform encephalopathies'], ['Bacterium', 'Negative', 'Vibrio cholerae', 'Cholera'], ['Bacterium', 'Negative', 'Shigella flexneri', 'Bacillary dysentery'], ['Bacterium', 'Negative', 'Mycobacterium tuberculosis', 'Tuberculosis'], ['Bacterium', 'Negative', 'Listeria monocytogenes', 'Listeriosis'], ['Bacterium', 'Negative', 'Helicobacter pylori', 'Peptic ulcers'], ['Bacterium', 'Negative', 'Chlamydia trachomatis', 'Chlamydial diseases']]Dimensions
A nested array is considered two dimensional or 2D when:
all of the members in a nested array are arrays themselves;
all of the sub-arrays have the same length — i.e. all the columns in the table are filled and have the same number of rows; and,
all of the members of the sub-arrays are homogeneous in type — i.e. they all have the same type (e.g.
int).
A two dimensional arrays may be visualised as follows: 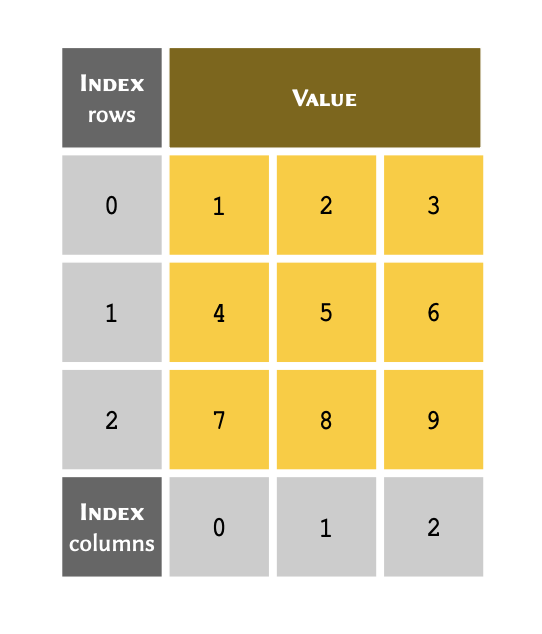
Advanced Topic
Nested arrays may themselves be nested. This means that, if needed, we can have 3, 4 or n dimensional arrays, too. Analysis and organisation of such arrays is an important part of a field known as optimisation in computer science and mathematics. Optimisation is itself the cornerstone of machine learning, and addresses the problem known as curse of dimensionality.
Such arrays are referred to in mathematics as a matrix. We can therefore represent a two-dimensional array as a mathematical matrix. To that end, the above array would translate to the annotation displayed in equation below.
\[table=\begin{bmatrix} 1&2&3\\ 4&5&6\\ 7&8&9\\ \end{bmatrix}\]
The implementation of these arrays is identical to the implementation of other nested arrays. We can therefore code our table in Python as:
OUTPUT
[[1, 2, 3], [4, 5, 6], [7, 8, 9]]OUTPUT
[7, 8, 9]OUTPUT
4OUTPUT
[[1, 2, 3], [4, 5, 6]]Do it Yourself
Computers see images as multidimensional arrays (matrices). In its simplest form, an image is a two-dimensional array containing only 2 colours.
Given the following black and white image:
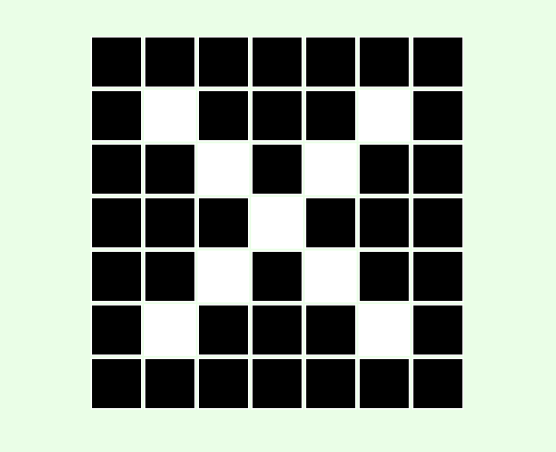
- Considering that black and white squares represent zeros and ones respectively, create a two-dimensional array to represent the above image. Display the results.
- Create a new array, but this time use
FalseandTrueto represent black and white respectively.
Display the results.
PYTHON
cross = [
[0, 0, 0, 0, 0, 0, 0],
[0, 1, 0, 0, 0, 1, 0],
[0, 0, 1, 0, 1, 0, 0],
[0, 0, 0, 1, 0, 0, 0],
[0, 0, 1, 0, 1, 0, 0],
[0, 1, 0, 0, 0, 1, 0],
[0, 0, 0, 0, 0, 0, 0]
]
print(cross)OUTPUT
[[0, 0, 0, 0, 0, 0, 0], [0, 1, 0, 0, 0, 1, 0], [0, 0, 1, 0, 1, 0, 0], [0, 0, 0, 1, 0, 0, 0], [0, 0, 1, 0, 1, 0, 0], [0, 1, 0, 0, 0, 1, 0], [0, 0, 0, 0, 0, 0, 0]]PYTHON
cross_bool = [
[False, False, False, False, False, False, False],
[False, True, False, False, False, True, False],
[False, False, True, False, True, False, False],
[False, False, False, True, False, False, False],
[False, False, True, False, True, False, False],
[False, True, False, False, False, True, False],
[False, False, False, False, False, False, False]
]
print(cross_bool)OUTPUT
[[False, False, False, False, False, False, False], [False, True, False, False, False, True, False], [False, False, True, False, True, False, False], [False, False, False, True, False, False, False], [False, False, True, False, True, False, False], [False, True, False, False, False, True, False], [False, False, False, False, False, False, False]]Summary
At this point, you should be familiar with arrays and how they work
in general. Throughout this section, we talked about list,
which is one the most popular types of built-in arrays in
Python. To that end, we learned:
how to
listfrom the scratch;how to manipulate
listusing different methods;how to use indexing and slicing techniques to our advantage;
mutability — a concept we revisit in the forthcoming lessons;
in-place operations, and the difference between weak references and deep copies;
nested and multi-dimensional arrays; and,
how to convert other sequences (e.g.
str) tolist.
Tuple
Data Structures: Tuples and Sequences
Another type of built-in arrays, tuple is an immutable
alternative to list. That is, once created, the contents
may not be modified in any way. One reason we use tuples is to ensure
that the contents of our array does not change accidentally.
For instance, we know that in the Wnt signaling pathway, there are two co-receptors. This is final, and would not change at any point in our programme.
OUTPUT
<class 'tuple'>OUTPUT
('Frizzled', 'LRP')OUTPUT
<class 'tuple'>OUTPUT
('Wnt Signaling', ('Frizzled', 'LRP'))OUTPUT
Wnt Signaling
Indexing and slicing principles for tuple is identical to
list, which we discussed in subsection indexing and slicing respectively.
Conversion to tuple
Similar to list, we can convert other sequences to
tuple:
OUTPUT
<class 'list'>OUTPUT
(1, 2, 3, 4, 5)OUTPUT
<class 'tuple'>OUTPUT
<class 'str'>OUTPUT
('T', 'h', 'i', 's', ' ', 'i', 's', ' ', 'a', ' ', 's', 't', 'r', 'i', 'n', 'g', '.')OUTPUT
<class 'tuple'>Immutability
In contrast with list, however, if we attempt to change
the contents of a tuple, a TypeError is
raised:
OUTPUT
TypeError: 'tuple' object does not support item assignmentEven though tuple is an immutable type, it can contain
both mutable and immutable objects:
PYTHON
# (immutable, immutable, immutable, mutable)
mixed_tuple = (1, 2.5, 'abc', (3, 4), [5, 6])
print(mixed_tuple)OUTPUT
(1, 2.5, 'abc', (3, 4), [5, 6])and mutable objects inside a tuple may still be
changed:
OUTPUT
(1, 2.5, 'abc', (3, 4), [5, 6]) <class 'tuple'>OUTPUT
[5, 6] <class 'list'>Advanced Topic
Why / how can we change mutable objects inside a
tuple when it is immutable? Members of a
tuple or not directly stored in the memory. An immutable
value (e.g. an int) has an existing, predefined
reference in the memory. When used in a tuple, it is that
reference that is associated with the tuple, and
not the value itself. On the other hand, a mutable object does not have
a predefined reference in the memory and is instead created on request
somewhere in the memory (wherever there is enough free space). Whilst we
can never change or redefine predefined references, we can always
manipulate something we have defined ourselves. When we make such an
alteration, the location of our mutable object in the memory may well
change, but its reference — which is what is stored in a
tuple, remains identical. You can find out what is the
reference an object in Python using the function id(). If
you experiment with it, you will notice that the reference to an
immutable object (e.g. an int value) would never
change, no matter how many time you define it in a different context or
variable. In contrast, the reference number to a mutable object
(e.g. a list) changes every time it is defined,
even if it contains exactly the same values.
OUTPUT
(1, 2.5, 'abc', (3, 4), [5, 15])OUTPUT
(1, 2.5, 'abc', (3, 4), [5, 15, 25])PYTHON
# We cannot remove the list from the tuple,
# but we can empty it by clearing its members:
mixed_tuple[4].clear()
print(mixed_tuple)OUTPUT
(1, 2.5, 'abc', (3, 4), [])Tuples may be empty or have a single value (singleton):
OUTPUT
() <class 'tuple'> 0PYTHON
# Empty parentheses also generate an empty tuple.
# Remember: we cannot add values to an empty tuple later.
member_b = ()
print(member_b, type(member_b), len(member_b))OUTPUT
() <class 'tuple'> 0PYTHON
# Singleton - Note that it is essential to include
# a comma after the value in a single-member tuple:
member_c = ('John Doe',)
print(member_c, type(member_c), len(member_c))OUTPUT
('John Doe',) <class 'tuple'> 1PYTHON
# If the comma is not included, a singleton tuple
# is not constructed:
member_d = ('John Doe')
print(member_d, type(member_d), len(member_d))OUTPUT
John Doe <class 'str'> 8Packing and unpacking
A tuple may be constructed without parenthesis. This is
an implicit operation and is known as packing.
OUTPUT
(1, 2, 3, 5, 7, 11) <class 'tuple'> 6PYTHON
# Note that for a singleton, we still need to
# include the comma.
member = 'John Doe',
print(member, type(member), len(member))OUTPUT
('John Doe',) <class 'tuple'> 1The reverse of this process is known as unpacking. Unpacking is no longer considered an implicit process because it replaces unnamed values inside an array, with named variables:
OUTPUT
14OUTPUT
14 17PYTHON
member = ('Jane Doe', 28, 'London', 'Student', 'Female')
name, age, city, status, gender = member
print('Name:', name, '- Age:', age)OUTPUT
Name: Jane Doe - Age: 28Note
There is another type of tuple in Python entitled
namedtuple. It allows for the members of a
tuple to be named independently (e.g.
member.name or member.age), and thereby
eliminates the need for unpacking. It was originally implemented by Raymond Hettinger, one of
Python’s core developers, for Python 2.4 (in 2004) but was much
neglected at the time. It has since gained popularity as a very useful
tool. namedtuple is not a built-in tool, so it is not
discussed here. However, it is included in the default library and is
installed as a part of Python. If you are particularly adventurous, or
want to learn more, feel free to have a look at the official
documentations and examples. Raymond is also a regular speaker at
PyCon (International Python Conferences), recordings of which are
available on YouTube. He also uses his Twitter to talk about small, but
important features in Python (yes, tweets!).
Summary
In this section, we learned about tuple, another type of
built-in arrays in Python that is immutable. This means that
once created, the array can no longer be altered. We saw that trying to
change the value of a tuple raises a
TypeError. We also established that list and
tuple follow an identical indexing protocol, and that they
have 2 methods in common: .index()() and .count().
Finally, we talked about packing and unpacking
techniques, and how they improve the quality and readability of our
code.
If you are interested in learning about list and
tuple in more depth, have a look at the official
documentations of Sequence Types – list, tuple, range.
Exercises
End of chapter Exercises
- We have
table = [[1, 2, 3], ['a', 'b'], [1.5, 'b', 4], [2]]what is the length of table and why?
Store your answer in a variable and display it using print().
- Given the sequence for the Gamma (catalytic) subunit of the Protein Kinase A as:
human_pka_gamma = (
'MAAPAAATAMGNAPAKKDTEQEESVNEFLAKARGDFLYRWGNPAQNTASSDQFERLRTLGMGSFGRVML'
'VRHQETGGHYAMKILNKQKVVKMKQVEHILNEKRILQAIDFPFLVKLQFSFKDNSYLYLVMEYVPGGEM'
'FSRLQRVGRFSEPHACFYAAQVVLAVQYLHSLDLIHRDLKPENLLIDQQGYLQVTDFGFAKRVKGRTWT'
'LCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAVGFPPFYADQPIQIYEKIVSGRVRFPSKLSSDLK'
'DLLRSLLQVDLTKRFGNLRNGVGDIKNHKWFATTSWIAIYEKKVEAPFIPKYTGPGDASNFDDYEEEEL'
'RISINEKCAKEFSEF'
)Using the sequence;
work out and display the number of Serine (S) residues.
work out and display the number of Threonine (T) residues.
calculate and display the total number of Serine and Threonine residues in the following format:
Serine: X
Threonine: X- create a nested array to represent the following table, and call it :
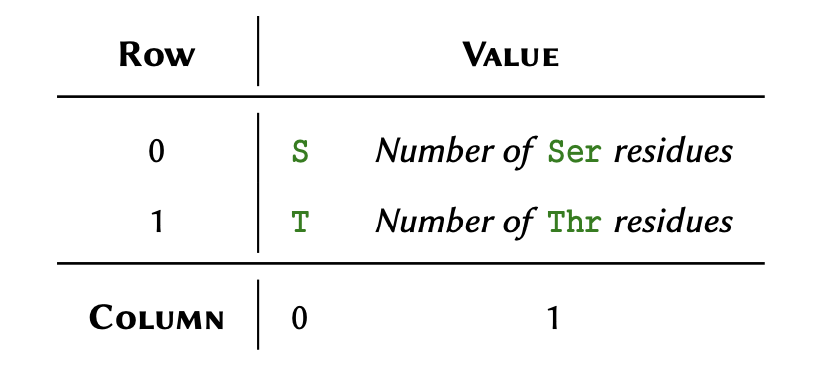
Explain why in the previous question, we used the term nested instead of two-dimensional in reference to the array? Store your answer in a variable and display it using print().
Graph theory is a prime object of discrete mathematics and is utilised for the non-linear analyses of data. The theory is extensively used in systems biology, and is gaining momentum in bioinformatics too. In essence, a graph is a structure that represents a set of object (nodes) and the connections between them (edges).
The aforementioned connections are described using a special binary (zero and one) matrix known as the adjacency matrix. The elements of this matrix indicate whether or not a pair of nodes in the graph are adjacent to one another.
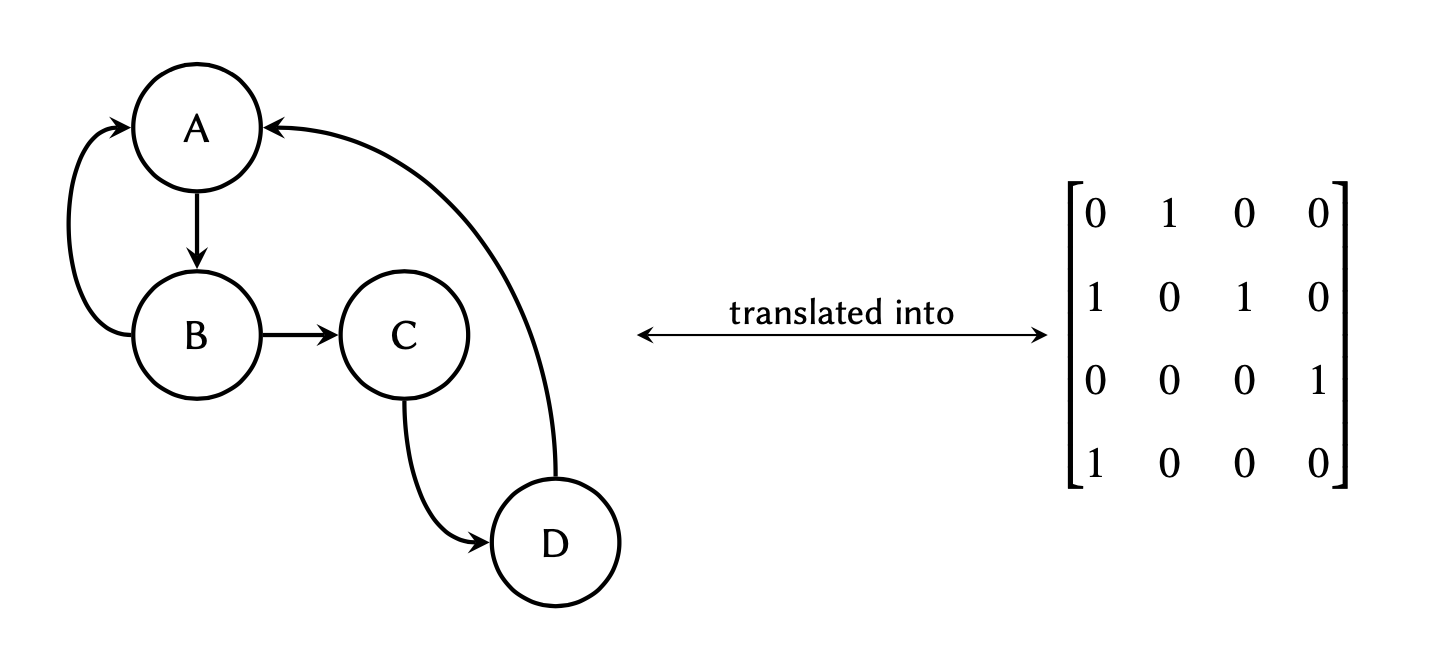 where each row in the matrix
represents a node of origin in the graph, and each column a node of
destination:
where each row in the matrix
represents a node of origin in the graph, and each column a node of
destination:
 If the graph maintains a
connection (edge) between 2 nodes (e.g. between nodes A and B in the graph above), the
corresponding value between those nodes would be #1 in the matrix, and
if there are no connections, the corresponding value would #0.
If the graph maintains a
connection (edge) between 2 nodes (e.g. between nodes A and B in the graph above), the
corresponding value between those nodes would be #1 in the matrix, and
if there are no connections, the corresponding value would #0.
Given the following graph: 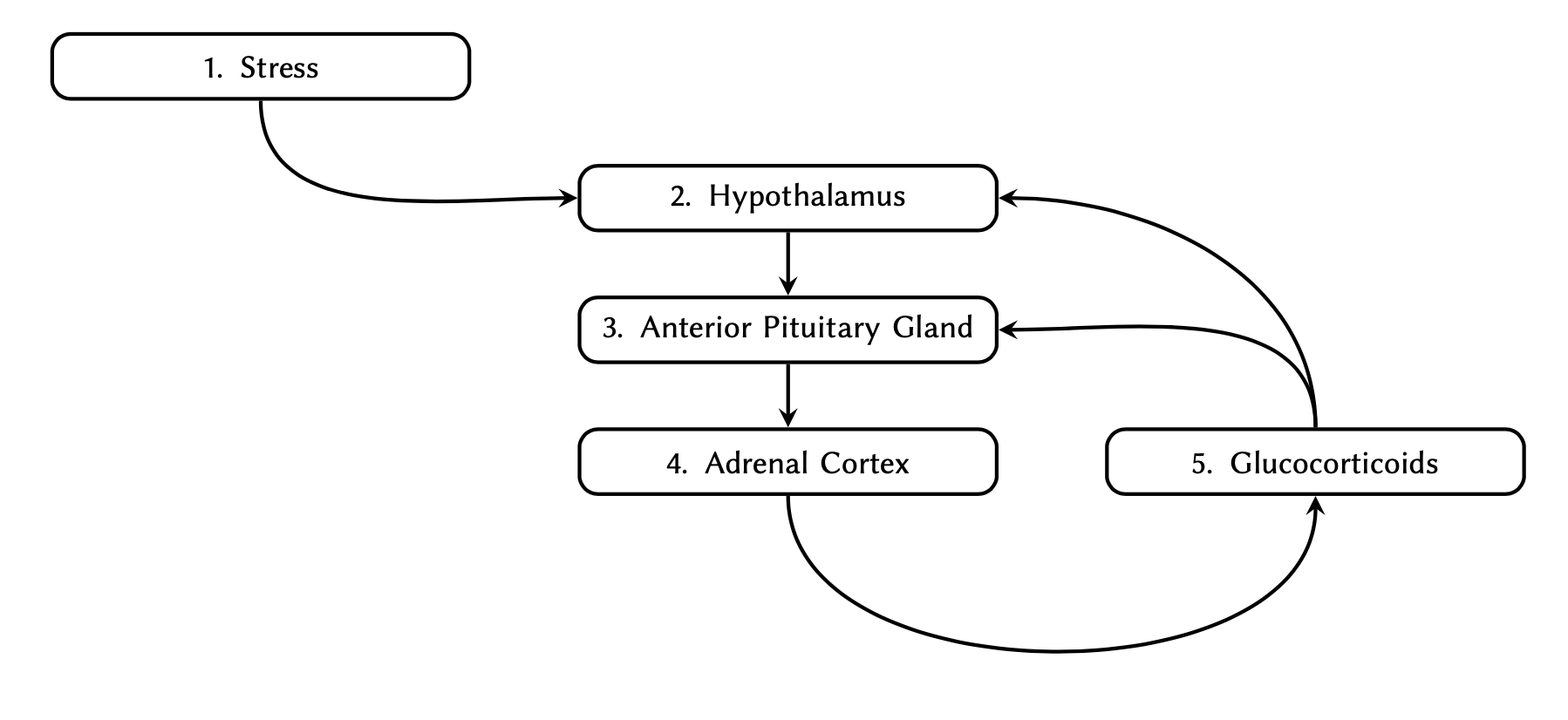
Determine the adjacency matrix and implement it as a two-dimensional array in Python. Display the final array.
Q1
PYTHON
table = [[1, 2, 3], ['a', 'b'], [1.5, 'b', 4], [2]]
table_length = len(table)
print('length of Table:', table_length)
reason = (
"The length of a `list` is a function of its "
"distinct members, regardless of their types."
)
print('')
print(reason)OUTPUT
length of Table: 4
The length of a `list` is a function of its distinct members, regardless of their types.Q2
PYTHON
human_pka_gamma = (
'MAAPAAATAMGNAPAKKDTEQEESVNEFLAKARGDFLYRWGNPAQNTASSDQFERLRTLGMGSFGRVML'
'VRHQETGGHYAMKILNKQKVVKMKQVEHILNEKRILQAIDFPFLVKLQFSFKDNSYLYLVMEYVPGGEM'
'FSRLQRVGRFSEPHACFYAAQVVLAVQYLHSLDLIHRDLKPENLLIDQQGYLQVTDFGFAKRVKGRTWT'
'LCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAVGFPPFYADQPIQIYEKIVSGRVRFPSKLSSDLK'
'DLLRSLLQVDLTKRFGNLRNGVGDIKNHKWFATTSWIAIYEKKVEAPFIPKYTGPGDASNFDDYEEEEL'
'RISINEKCAKEFSEF'
)
total_serine = human_pka_gamma.count("S")
total_threonine = human_pka_gamma.count("T")
print('Serine:', total_serine)
print('Threonine:', total_threonine)
residues = [
['S', total_serine],
['T', total_threonine]
]
print(residues)OUTPUT
Serine: 19
Threonine: 13
[['S', 19], ['T', 13]]Q3
PYTHON
answer = (
"Members of a two-dimensional array must themselves be arrays of "
"equal lengths containing identically typed members."
)
print(answer)OUTPUT
Members of a two-dimensional array must themselves be arrays of equal lengths containing identically typed members.Q4
PYTHON
# Column initials:
# S, H, A, A, G
adjacency_matrix = [
[0, 1, 0, 0, 0], # Stress
[0, 0, 1, 0, 0], # Hypothalamus
[0, 0, 0, 1, 0], # Anterior Pituitary Gland
[0, 0, 0, 0, 1], # Adrenal Cortex
[0, 1, 1, 0, 0], # Glucocorticoids
]
print(adjacency_matrix)OUTPUT
[[0, 1, 0, 0, 0], [0, 0, 1, 0, 0], [0, 0, 0, 1, 0], [0, 0, 0, 0, 1], [0, 1, 1, 0, 0]]Key Points
-
listsandtuplesare 2 types of arrays. - An index is a unique reference to a specific value and Python uses a zero-based indexing system.
-
listsare mutable because their contents to be modified. - slice(), .pop(), .index(), .remove() and .insert() are some of the key functions used on mutable arrays.
-
tuplesare immutable which means its contents cannot be modified.
